Data Practices for Open Science
Data Perils We’ve All Dealt With
Uh-oh!
- Which version of the dataset was the final one again?
- Who was the last person to edit the pipeline?
- Opening my dataset is slowing my computer down.
- Reviewer 2 wants changes. I need to repeat the entire analysis!
About Me
- Albury C
- MSc, Marine microbial trace nutrient dynamics, Dalhousie University
- Currently a data scientist for Infrastruture Canada

About You

“If I have seen further, it is by standing on the shoulders of giants.” - Sir Isaac Newton
Open Science
- Aims to make research accessible for the benefits of both scientists and broader society (UNESCO 2023)
- Increases transparency and the speed of knowledge transfer
FAIR Principles
- Findable
- Accessible
- Interoperable
- Reusable
FAIR Principles
- “Importantly, it is our intent that the principles apply not only to ‘data’ in the conventional sense, but also to the algorithms, tools, and workflows that led to that data.” (Wilkinson et al. 2016)
- Focus on machine actionability

Best Data Practices
- Documentation
- Storing Your Data
- Coding Practices
Documentation
- Use version control
- Master the art of the README
- Anticipate package conflicts
Documentation: Use version control
- Git is a popular SCM (source code manager). It can be used through your terminal, browser (ex: Github), or desktop software (ex: GitKraken or your chosen IDE).
- Version control helps you to avoid circumstances that birth monstrosities like
analysis_draft_3_final.R. - An example of a Github repository
Documentation: Use version control
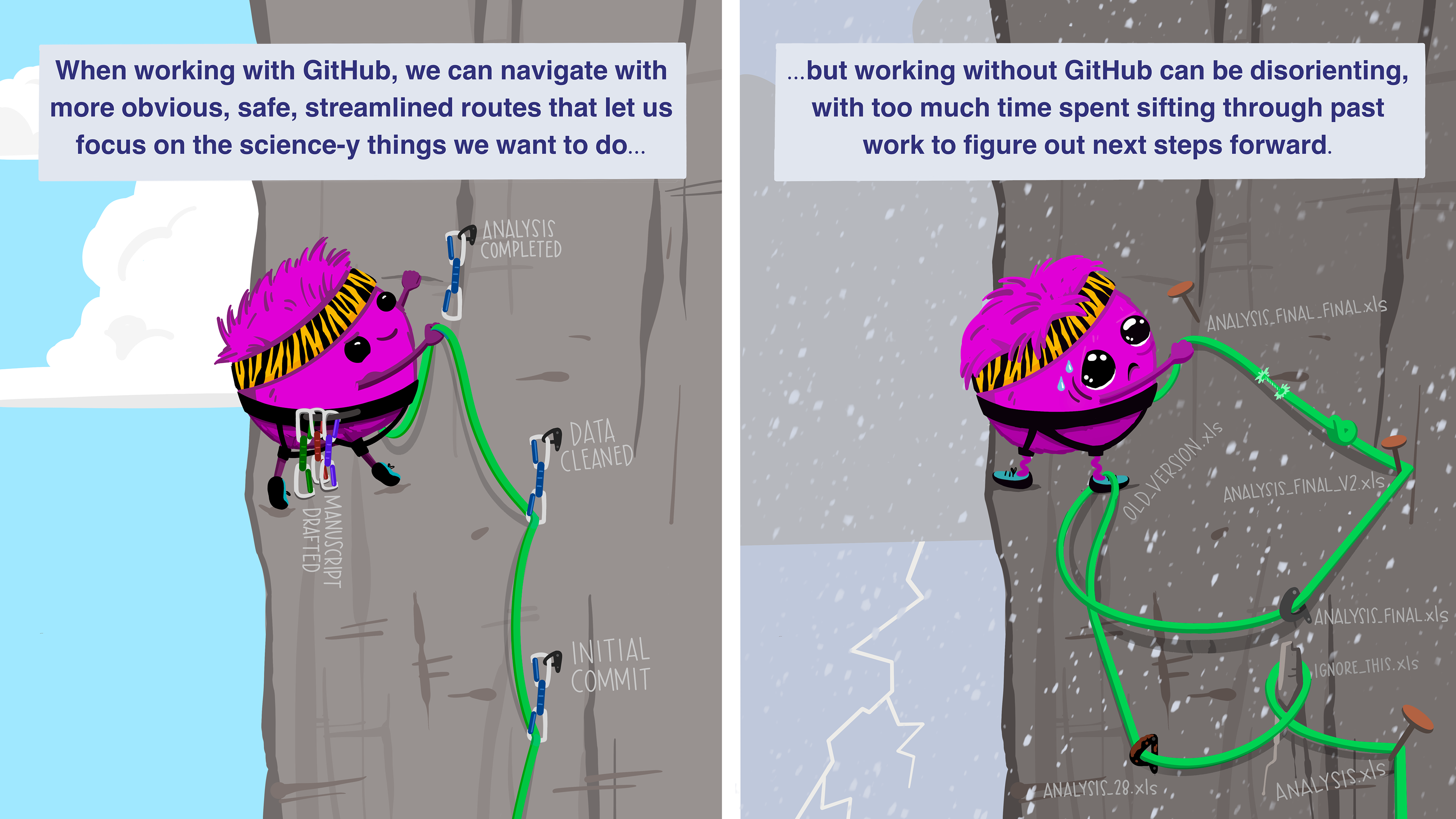
Illustration by Allison Horst
Documentation: Master the art of the README
- A README acts at the title page of your repository, letting folks who are knew the who, what, where, and why of the analysis.
- Use the README to document anything that might be important if someone brand new was to try to pick up where you’ve left off.
Documentation: Anticipate package conflicts
- Note the version of critical packages you used in your analysis. This helps with troubleshooting down the line.
- For advanced models, some use conda to manage environments and package versions.
Documentation: Anticipate package conflicts
Conflicted?
Many of us have had the experience of loading a package that contains a function with the same name as one our code relies on.
Use the conflicted package to choose which to use.
Storing Your Data
- Use tidy data format
- Redundancy in data is good
- Keep raw data raw
- Manage your memory
Storing Your Data: Use tidy data format
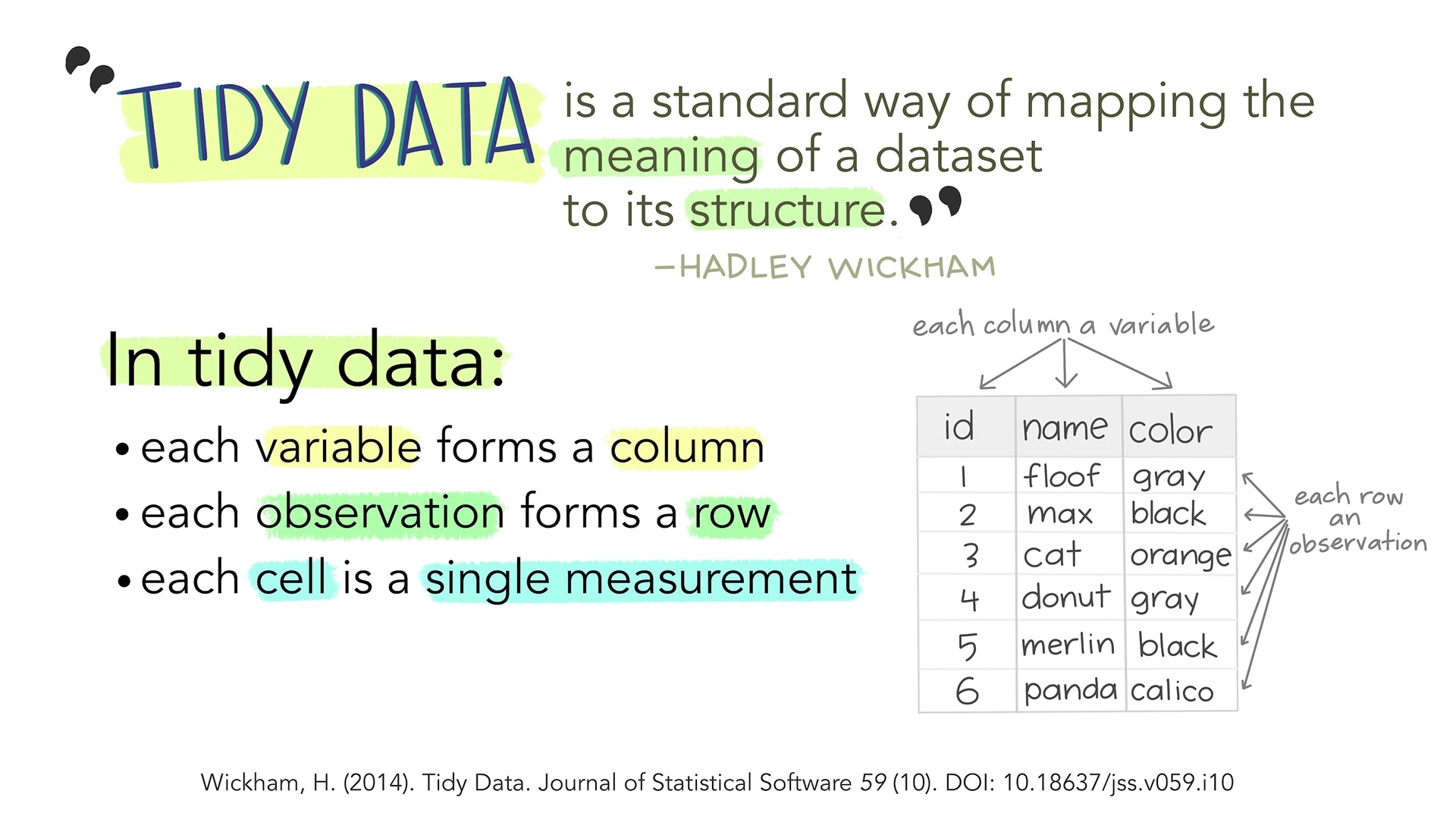
Illustration from Tidy Data for reproducibility, efficiency, and collaboration by Julia Lowndes and Allison Horst
Storing Your Data: Redundancy in data is good
- Utilize metadata and data dictionaries to link provide important context to your primary data set.
- Before beginning a project, think about the different ways that data should be stored.
- Ex: In a field study, observations from each study site should be kept separate from information about each study site. The two can be linked by unique site identifiers.
Storing Your Data: Redundancy in data is good
- This table contains two types of observations: song information and Billboard ranking (Wickam 2014)
- One entry each week the song remains on the Billboard top 100
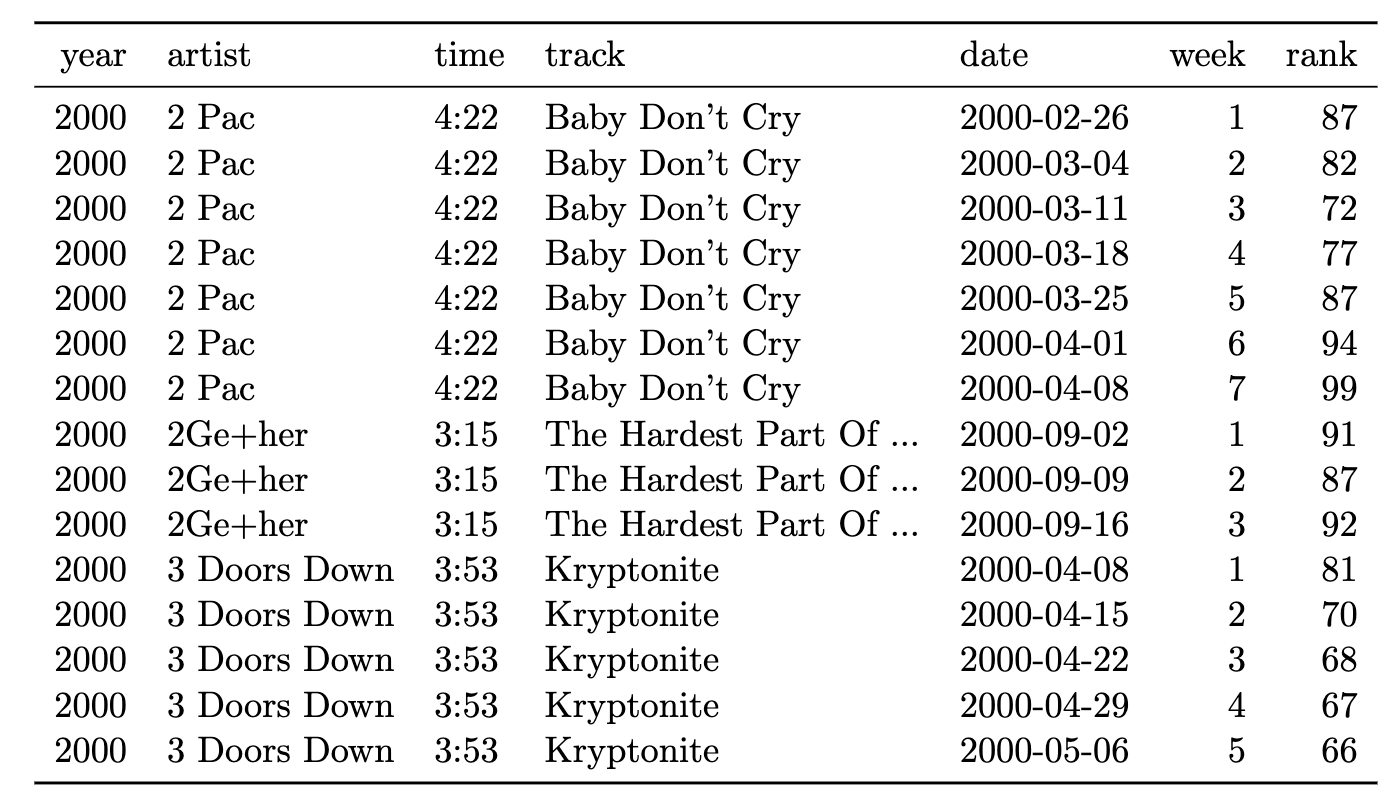
Storing Your Data: Redundancy in data is good
Instead, they should be saved as:
- a table with song’s artists, names and run times
- a table with details on their Billboard ranking (Wickam 2014).
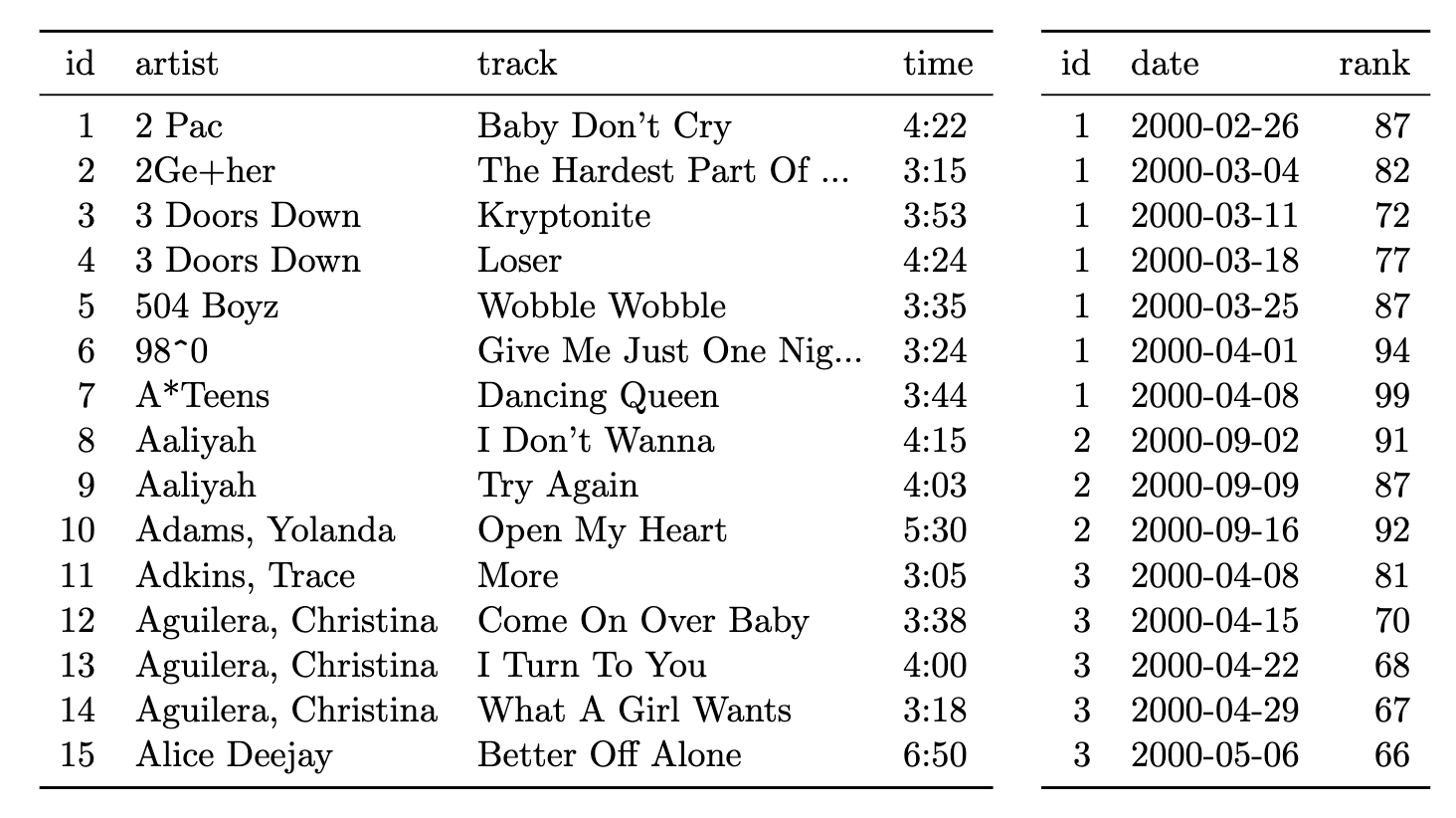
Storing Your Data: Redundancy in data is good
- Reduces each table to only one type of observation, avoiding confusion
- Saves space

Storing Your Data: Keep raw data raw
- In repositories, create a folder named RAW. Place collected data here.
- Do not open these files except to add or remove raw data. Consider setting them to read only when viewing in excel.
- Keep all analysis to your scripts, which will read the data and create outputs from it.
Storing Your Data: Manage your memory
- Keep data types in mind
- Consider parquet for large files
- Use the
arrowpackage to work with larger than memory data sets
Storing Your Data: Manage your memory
Coding Practices
- Follow a style guide
- Use relative file paths
- Avoid hard coded values
Coding Practices: Follow a style guide
The Tidyverse Style Guide is a great resource for style guidance.
Coding Practices: Follow a style guide
Auto-formatting
Use cmd + shift + A in R-studio to auto-format your code on the fly.
Coding Practices: More on Style
Sectioning your code makes it easy for you and others to find the part of the analysis that you’re looking for at a glance.
Auto-formatting
Use cmd + shift + R in Rstudio to create a new collapsible section.
Coding Practices: Use relative file paths
- Ever tried to run a collaborator’s script only to come up come up with
cannot open file 'important-data.csv': No such file or directory? They probably didn’t set the directory for you! - Don’t instruct your colleague’s computer to look for
Desktop/Medical_files/hair-transplant/important-data.csv. Be kind, use a relative file path instead.
Coding Practices: Use relative file paths
Relative paths specify locations starting from the current location.
Coding Practices: Use relative file paths
- Change directories throughout the script to access different folders in the repository.
# Relative file path to a dedicated folder with raw data files
setwd("~/example-repo/RAW")
read.csv("important-data.csv")
# Call on another R source file back at the root of the directory
setwd(".")
source("analysis.R")
# Save the results of the analysis in it's own folder
setwd("/Cleaned_data")
write.csv(data_output, "data_output.csv")Coding Practices: Avoid hard coded values
- Ever seen code where the logic is impossible to follow due to manual manipulations?
- If using constants, make sure they’re carefully commented.
- Otherwise, try to utilize variables whenever possible to improve reproducibility
Coding Practices: Avoid hard coded values
# Bad
spec_data |>
mutate(scatter_normalized = scatter / 2.8)
# Better
spec_data |>
# Normalize by dividing by constant from calibration
mutate(scatter_normalized = scatter / 2.8)
# Best
# Load analysis that generates constant
source("spec_calibration.R")
spec_data |>
# Normalize by dividing by constant from calibration
mutate(scatter_normalized = scatter / calibration_value)Conclusions
- There are many opportunities to modify your research workflows to both:
- Save your future self some headache
- Make your science more useful to others
- A list of resources can be found at the end of the presentation
Happy Data Wrangling!
References
- Wickham, H. (2014). Tidy Data. Journal of Statistical Software, 59(10), 1–23. https://doi.org/10.18637/jss.v059.i10
- Wilkinson, M., Dumontier, M., Aalbersberg, I. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3, 160018 (2016). https://doi.org/10.1038/sdata.2016.18
- UNESCO Recommendation on Open Science. 2023
Resources
- Software Carpentry: Version Control with Git
- Making README’s
- Tidyverse Style Guide
- Conflicted
- Large Data in R: Tools and Techniques
- Data Types in Arrow and R
- What is the Parquet File Format? Use Cases & Benefits
- R for Reproducible Research:Navigating Files and Directories
Data Practices for Open Science